PAG stands for Pathway, Annotated-list and Gene-signature. A PAG geneset represents a set of genes that share a common feature, whether they belong within the same pathway, share common biological processes, are co-differentially expressed under a particular condition of an experiment or clinical sample, or share a common downstream target or function. Each P-type PAG refers to a connected set of molecules (genes/proteins/metabolites), among which some detail of curated mechanism of actions, e.g. protein interactions, reactions, or gene regulations, are available (Biological Pathway, http://www.genome.gov/27530687). Each A-type PAG refers to a curated list of genes/proteins identified from a specific biological context, e.g. a shared GO category or a shared protein family. Each G-type PAG refers to a list of genes/proteins derived from any given high throughput Omics experiment, e.g. functional genomics, under a shared biological condition.
Pathway and Annotated-list Gene-signature Electronic Repository for Coronavirus-related disease (PAGER-CoV) is an online systems biology tool for constructing and visualizing gene and PAG networks from multiple PAG genesets.
PAGER-CoV contains domain-expert curated PAGs curated from published high-impact SARS-CoV-2 studies as well as curated PAGs relating to infectious disease inherited from PAGER 2.0. PAGER-CoV was designed specifically for COVID-19 drug repositioning and translational research studies, as well as to help COVID-19 researchers quickly identify relevant information based on a queried geneset. PAGs from PAGER-CoV can be used with GSEA software for parallel analysis.
PAGER was implemented by using PHP language version 5 and Codeigniter version 2.1.3. PAGER data was stored in Oracle 12g database maintained by Indiana University and was connected to PHP server by Oracle Instant Client software. P-value for each edge in PAGs' networks was computed on the fly by using hypergeometric function provided by PDL. For gene and PAGs' networks visualization in PAGER, cytoscape.js, a JavaScript graph library for network visualization.
PAGER-CoV currently contains a total of 3263 PAGs from 27 data sources, 281 of these PAGs being manually curated by a domain expert, and is being updated weekly. We welcome you to add your COVID-19-related PAGs to PAGER-CoV ( link ).
For full breakdown on PAG sources, refer to the table below:
| Category | Source or Journal | Description | Count | |
|---|---|---|---|---|
| PubChem | PubChem | A compiled list of PubChem data related to COVID-19 (Coronavirus Disease 2019) and SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) | 1549 | 1549 |
| PAGER | PAGER-MSigDB | Immune signatures from GSEA (imported from PAGER 2.0). | 6139 | 10015 |
| PAGER-GOA_AGG | Gene Ontology Annotation database (is_a relation aggregation). | 1888 | ||
| PAGER-GAD | Gene signatures from Genetic Association Database. | 70 | ||
| PAGER-GOA | Gene Ontology Annotation database. | 1030 | ||
| PAGER-Pfam | A Large collection of protein families represented by multiple sequence alignments and hidden Markov models (HMMs). | 82 | ||
| PAGER-GeneSigDB | A curated database of gene expression signatures. | 390 | ||
| PAGER-Protein Lounge | Interactive web-based biological database. | 30 | ||
| PAGER-Spike | Signaling Pathways Integrated Knowledge Engine, regulation from highly curated human pathway information. | 3 | ||
| PAGER-PheWAS | Phenome-wide association studies database. | 57 | ||
| PAGER-PharmGKB | A database containing information relating to human genetic variation on drug response. | 4 | ||
| PAGER-Reactome | An interactive pathway database. | 25 | ||
| PAGER-BioCarta | Online maps of metabolic and signaling pathways. | 105 | ||
| PAGER-GWAS Catalog | GWAS Catalog A Catalog of Published Genome-Wide Association Studies provided by NHGRI | 79 | ||
| PAGER-KEGG | A database containing high-level functions and utilities of the biological system. | 38 | ||
| PAGER-NGS Catalog | A Database of Next Generation Sequencing Studies in Humans. | 1 | ||
| PAGER-DSigDB | A drug signatures database for gene set analysis | 49 | ||
| PAGER-GTEx | The Genotype-Tissue Expression (GTEx) project | 2 | ||
| PAGER-NCI-Nature Curated | Gathered from published research literature and reviewed by expert scientists before publication. | 13 | ||
| PAGER-WikiPathway | A database of biological pathways maintained by and for the scientific community. | 10 | ||
| Curation | Am J Respir Crit Care Med | Clinical journals related to pulmonary, critical care, and sleep-related fields. | 2 | 271 |
| Microbiology and Molecular Biology Reviews | Research journals related to immunology, molecular, and cell biology. | 1 | ||
| Zenodo | general-purpose open-access repository developed under the European OpenAIRE program and operated by CERN. | 1 | ||
| Mouse Genome Informatics Database | international database resource for the laboratory mouse, providing integrated genetic, genomic, and biological data for researching human health. | 5 | ||
| Nature Cell Discovery | Cell molecular biology journal. | 4 | ||
| GenBank/UniProt | Databases used to manually curate gene to protein mapping and protein annotations. | 33 | ||
| Cell | A research journal relating to cell biology | 5 | ||
| The Annual Review of Cell and Developmental Biology | A research journal relating to developments in the field of cell and developmental biology. | 1 | ||
| Nature | A leading multidisciplinary science journal. | 111 | ||
| Drugbank | Unique bioinformatics and cheminformatics resource that combines detailed drug data with comprehensive drug target information. | 96 | ||
| Nature Medicine | A monthly journal publishing original peer-reviewed research in all areas of medicine. | 1 | ||
| Cell Host and Microbe | A research journal that focuses broadly on the study of microbes, with an emphasis on the interface between the microbe and its host. | 1 | ||
| bioRxiv | COVID-19 SARS-CoV-2 preprints from medRxiv and bioRxiv. | 10 | ||
| Total | 11835 | |||
PAGER-CoV has several unique features available:
- PAGER not only allows users to construct two types of PAG networks, PAGER enables users to construct gene interaction or gene regulation networks within a PAG.
- PAGER allows users to construct three types of expanded PAG networks including upstream, downstream, and co-membership PAGs. Constructing expanded PAGs networks enables users to find key genesets related to their study.
- Because PAGER-CoV offers gene networks, users can also construct expanded gene networks from a gene including networks of upstream, downstream, and sibling genes.
- PAGER provides an interactive visualization tool for users to study gene and PAGs' networks and offers spaces, Gene Box and PAG Box, for users to store their genes and PAGs.
For examples of how to use PAGER, please see Use Cases.
Users can perform a basic search query for PAGs in PAGER-CoV by entering a keyword or gene symbol into the search bar on the home page:
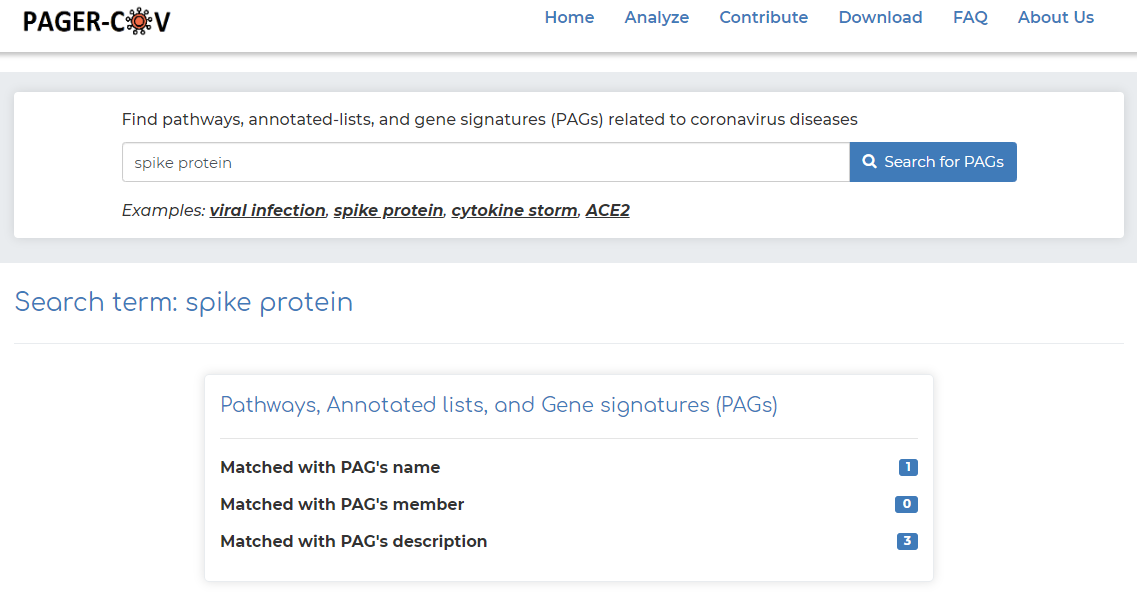
- As an example, click on ‘spike protein’ example under the basic search bar and click ‘Search for PAGs’.
- After searching, we see that PAGER-CoV brings a total of 4 results, with the ‘spike protein’ search query matching with 1 PAG name, and 3 PAG descriptions.
- Clicking on the blue box to the right of PAGER-CoV results (in our example we click the blue box representing the PAG name matched with ‘spike protein’ query), we get:
- PAG summary information with reference to the original data source:
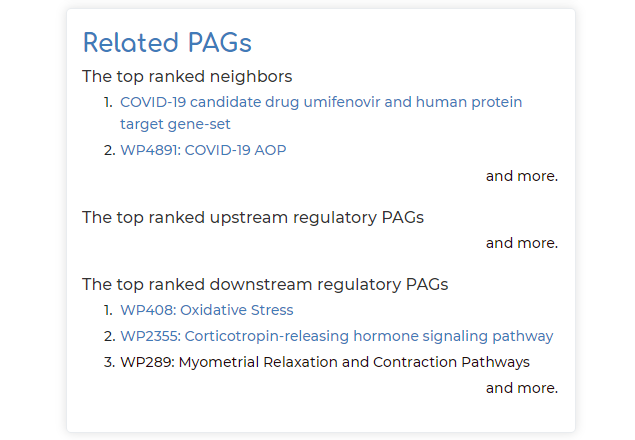
- Top-ranked related PAG neighbors, as well as top-ranked regulatory PAGs that are upstream or downstream to PAG of interest (note in our example, a basic ‘spike protein’ search query brings up the top-ranked PAG neighbor as a PAG relating to COVID-19 candidate drug umifenovir). These related PAGs can be clicked on if users are interested to explore further:
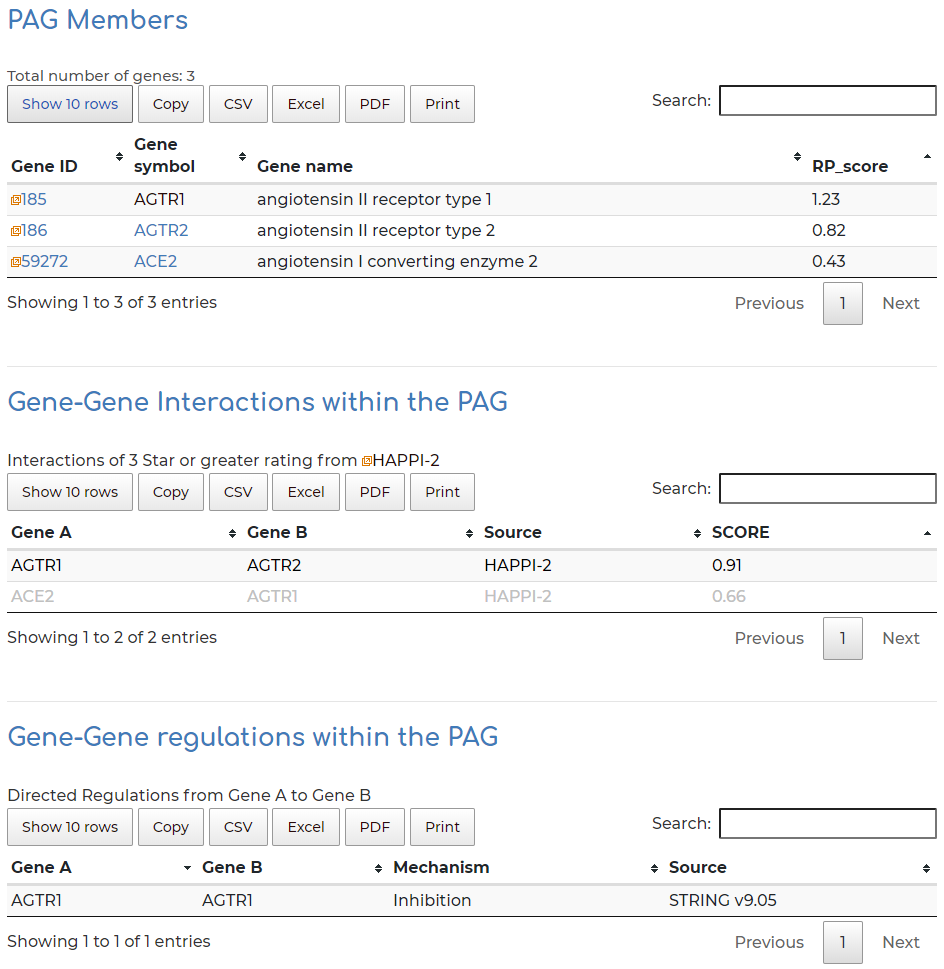
- Gene members within the PAG, alongside PAG gene member interaction and regulatory information:
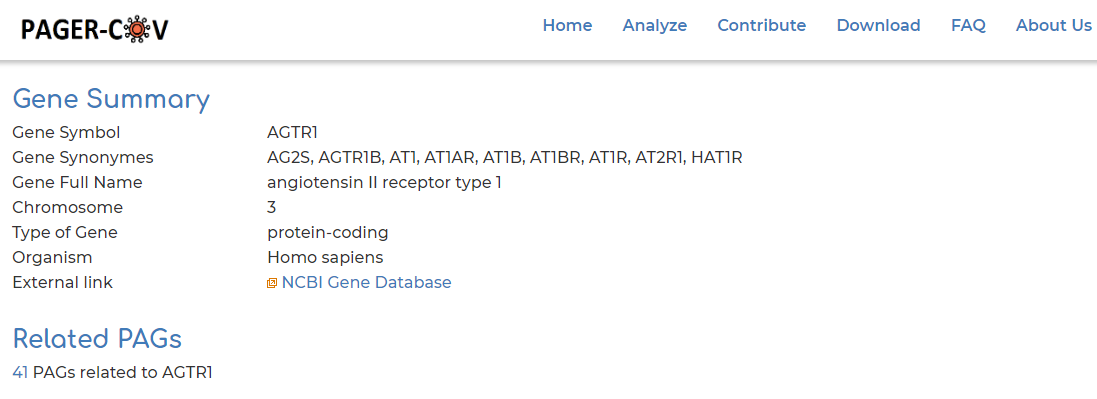
- Users can click on ‘Gene symbol’ within ‘PAG Members’ section to access information to PAG gene information. In our example we show the AGTR2 gene:
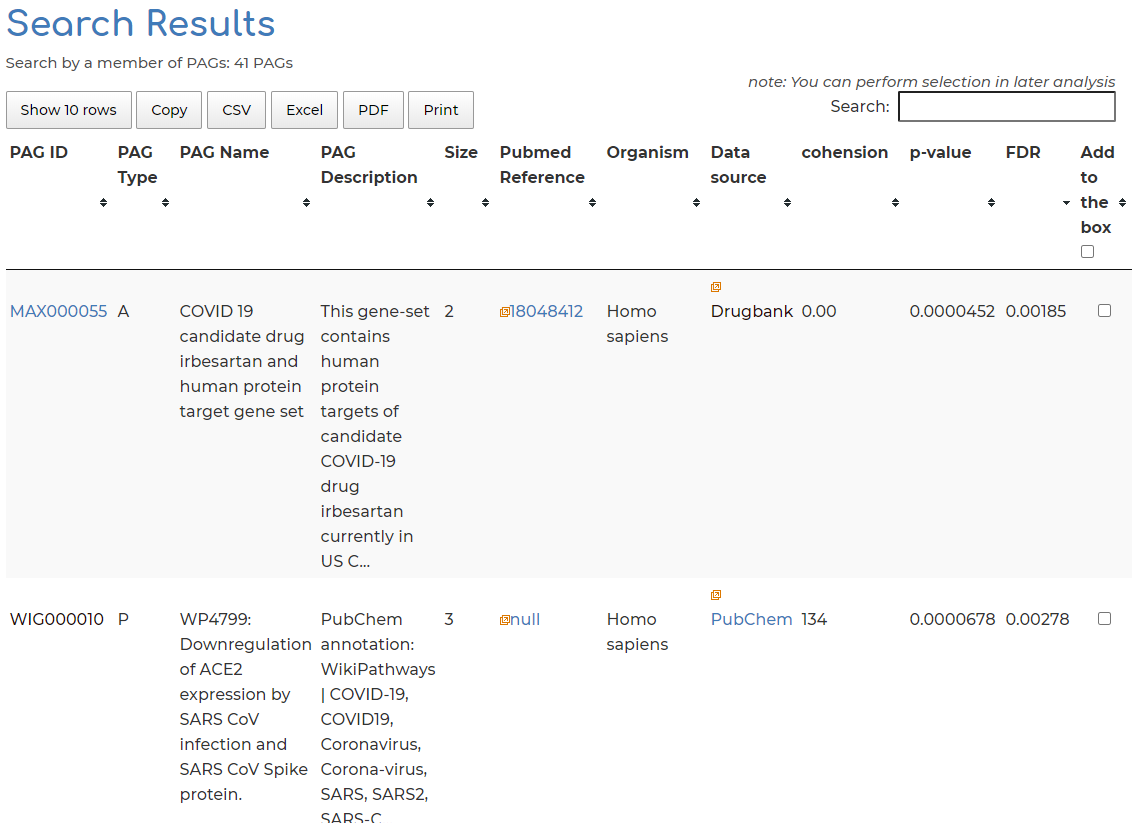
- Clicking on ‘Related PAGs’ section from Step 4, we can see the 33 PAGs which AGTR2 is a member of in PAGER-CoV:

Users can enter a list of genes obtained from their experiment or other data source to order to search for related PAGs in PAGER-CoV:
- On the PAGER-CoV home page, click on 'Advanced Search' option.
- Clicking on the example gene set provided in PAGER-CoV: 'COVID-19 clinical samples mild vs. severe', representing 1516 differentially expressed genes in patients with mild vs. severe cases of COVID-19.
- Click 'Search PAGs'.
- After searching, we see 49 PAGs similar to our queried geneset, based on the standard filters. Users can view PAG results by clicking on the header row of table, based on overlap of number of genes (default setting), similarity score between query gene set and hits, CoCo score, p-value, etc:
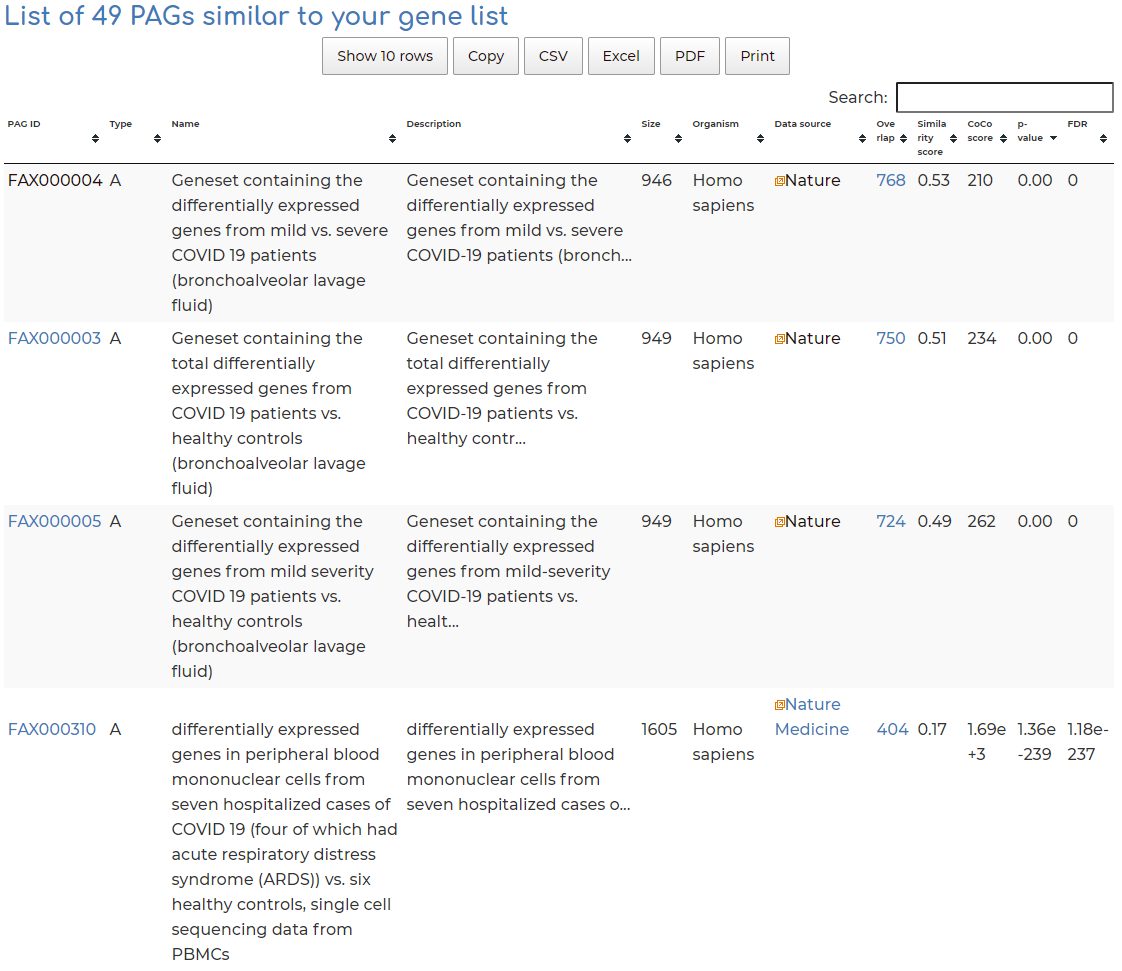
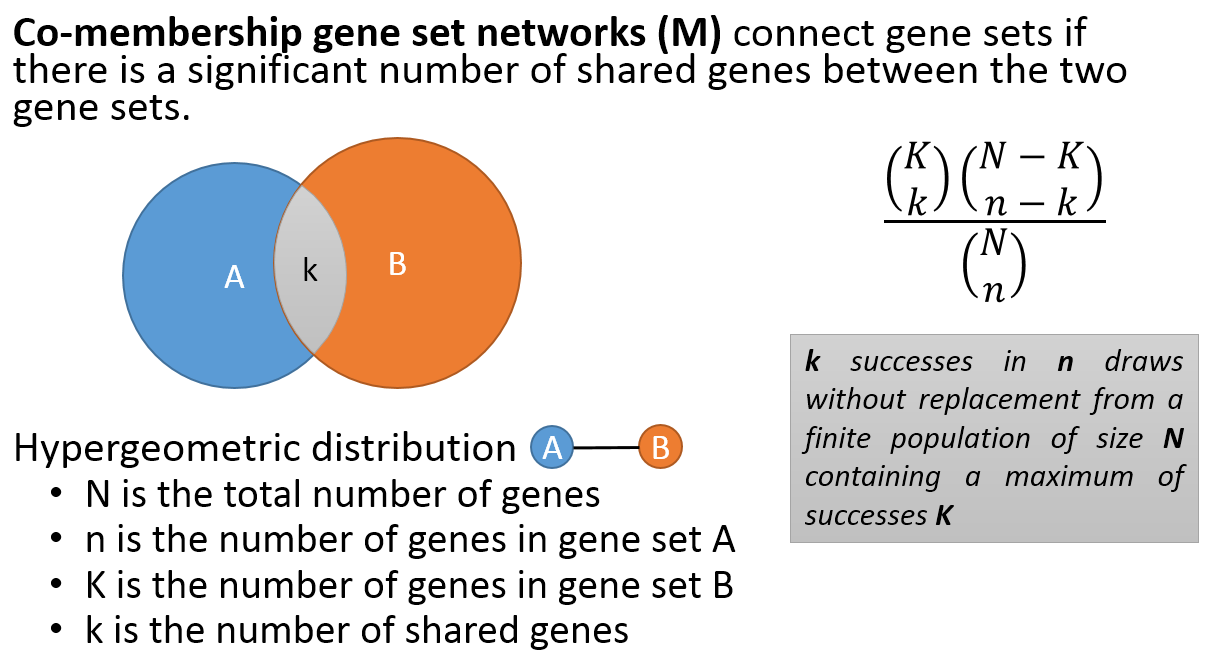
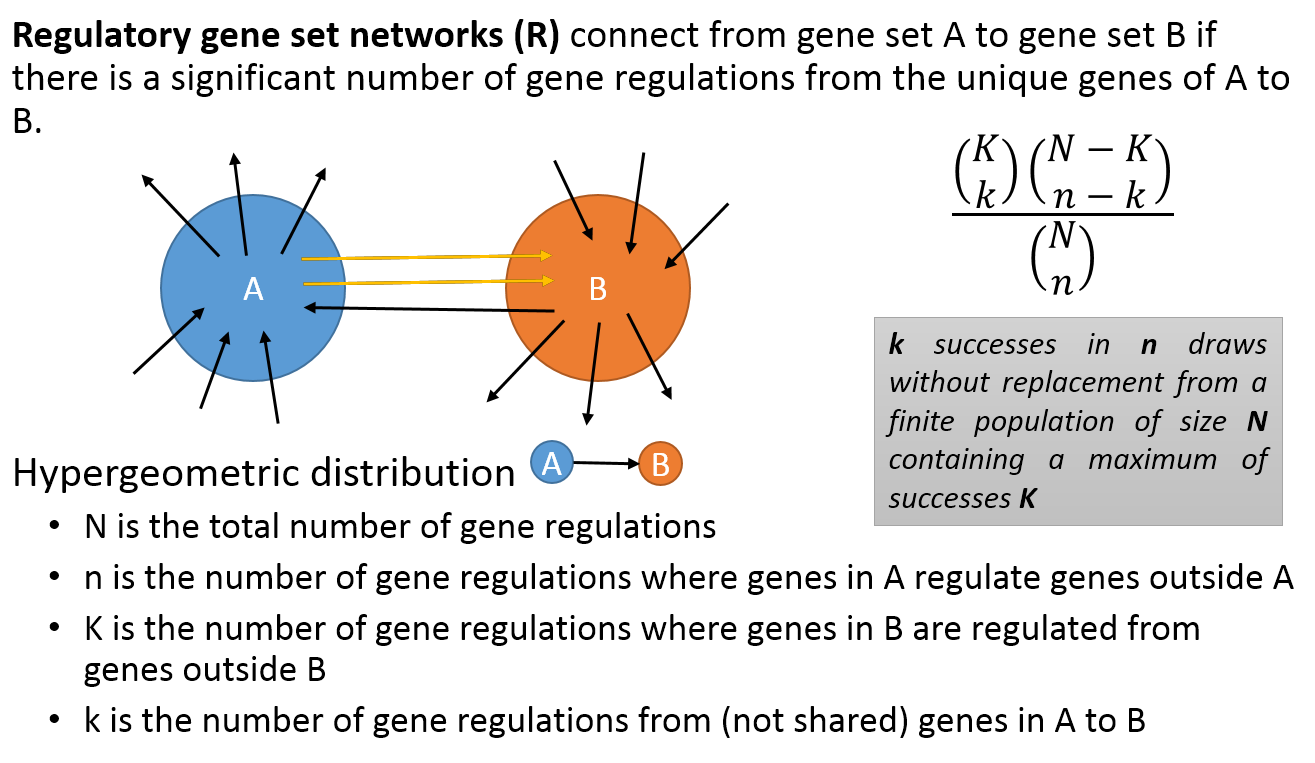
- Co-membership PAG-to-PAG networks (m-type PAG-to-PAG networks) connect a pair of PAGs if there is a significant number of shared genes.
- A regulatory PAG-to-PAG network (r-type PAG-to-PAG network) connects a pair of PAGs when there is a significant number of gene regulations between the genes of the two PAGs.
The RP-score (standing for rank prioritization-score) is used to rank genes within the PAG, allowing for output of a PAGER-ranked gene list.
We use RP-score to calculate and assign the gene weight in every PAG:
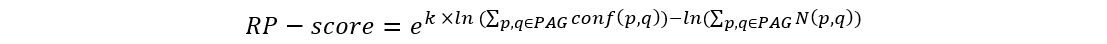
where p and q are the indexes of proteins from the selected module, k is a constant (k=2 in this study). The term conf(p, q) is the interaction confidence score assigned by HAPPI-2, where conf(p, q) is between 0 and 1. N(p, q) holds the value of 1 if protein p interacts with q or the value of 0 if protein p does not interact with q. We then rank the genes by the RP-score to pull out potential important genes in the top rank list.
The nCoCo score takes into account PAG size bias when determining quality score of PAG results. Users can view PAGs similar to their queried gene list by nCoCo score by clicking on header arrows:
The nCoCo score represents a PAG quality metric and is improved from the initial CoCo score (a Cohesion Coefficient derived from the measure of statistically significant coverage of gene-gene functional correlations in gene pairs or gene trios) in our PAGER publication .
Yes! Users may refer to this link for detailed instructions.
Yes! Users may refer to this link for detailed instructions.
The PAGER-CoV API allows developers to analyze gene lists, get enrichment results, and download files of enrichment results. We provide a demo using two R scripts in performing the PAGER-CoV analysis using API.
-
The core function script for accessing the PAGER-CoV API:
- PAGERCOV.r The script of calling the functions:
- PAGERCOV_API_CALL.r